Genomics, the study of genetic information, focuses on analyzing the functionalities, spatial structure and evolution of genomes. It is an extensive area of study, rendered more complex by the amorphous nature of genomes and genetic information encoding in many species.
In the 21st-century, genomics is also the bedrock for the extraction of information regarding the evolution of species, profiling of hereditary diseases, and the development of individual-specific diagnosis and medication. Scientific advancements are unveiling new possibilities every day in genomics and transforming biomedical disciplines such as anthropology, paleontology, healthcare and life sciences. Development of data analytics and GPU-accelerated computation has further empowered genome analytics to deliver stupendous precision at unprecedented speeds.
Let us learn more about what genomics is, the challenges associated with genome sequencing, and some of its use cases. We will also delve into how GPUs and cloud computing can accelerate next-generation genome sequencing compared to CPUs.
What is Genome Sequencing?
Genome sequencing refers to the compilation of the DNA sequences of individual organisms to aid in disease research, vaccine development and tracing the evolutionary origins of a species/ community. In-depth analysis of genome sequences can reveal novel insights and help address biomedical/ evolutionary challenges. Genome sequencing has witnessed immense technology advancements in recent years, both in terms of hardware and software-based solutions, and several advanced sequencing systems are available off-the-shelf in the market now for healthcare establishments, research institutions and medical enterprises.
There have been several instances where genome analysis has been in the news recently –
- Ongoing research into mammalian genomes and the discovery of over three million regulatory elements: These insights added to the understanding of mammalian evolution, disease tendencies, species-specific traits and extinction risks.
- Identification of genetic factors leading to common medical conditions such as diabetes, high blood pressure and cancer.
- Contribution to the understanding of monogenic disorders, including the introduction of preventive medication/ clinical care for individuals carrying disease-causing genes prior to the onset of high-risk diseases.
- Personalized medication and interventions tailored to individual needs.
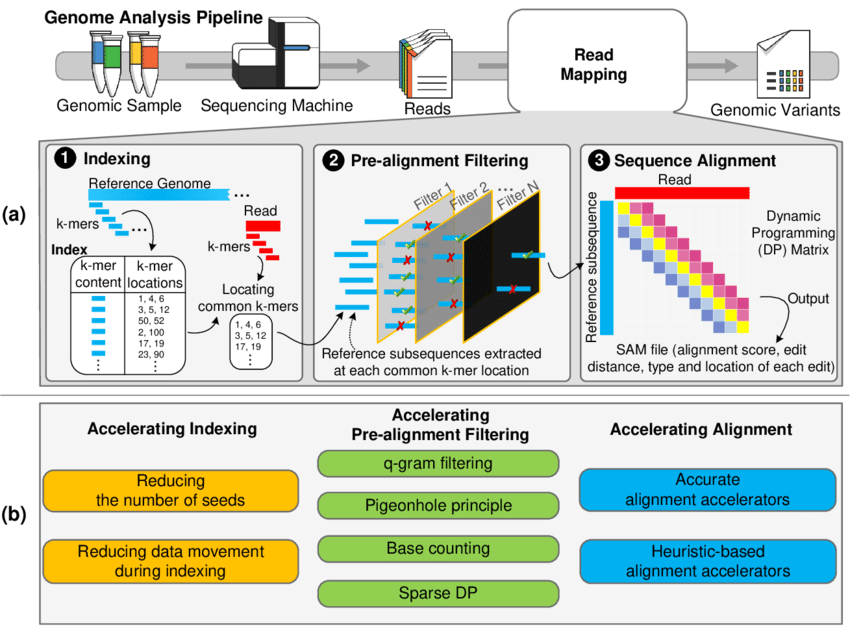
Approaches to accelerate various steps of read mapping process in genome analytics (Source)
What are the Challenges of Genome Analysis?
Like most data analytics, genome analysis also involves handling voluminous information, factoring-in extensive variables, and performing computationally intensive calculations. There are other domain-specific challenges as well. For instance, if we focus on healthcare/ clinical genomics, key bottlenecks are –
- Expert researchers: Integrating genomics with other disciplines, like virology or evolutionary biology, demands a diverse team of specialists like research analysts and bioinformatics engineers, etc. Naturally, such projects incur significant expenditure, especially in terms of co-location of all the necessary resources and manpower.
- Excess information: Despite the potential of genome analysis to advance precision medicine, healthcare providers must exercise extreme caution in sharing diagnoses with patients as the latter may struggle to cope with information that lacks solid analytical and observational foundations.
- Confusing data: While genomic data can provide valuable insights into patient health, the sheer volume of data can be overwhelming, and researchers may struggle to accurately discern which information is relevant to their domain/ study at hand.
- Inadequate infrastructure: Challenges pertaining to genomic data management, infrastructure modernization and data integration are primary obstacles faced by healthcare firms in their quest to incorporate genomic analytics in clinical care and diagnosis.
Also Read: GPU Vs. CPU For Image Processing: Which One Is Better?
Shift Your Genomic Research to Cloud Computing Now!
Genome analytics, gene sequencing and genetic engineering have made significant advancements in recent years. Gene research firms and healthcare organizations have deftly integrated their operations with cloud computing which has significantly reduced the cost of genomic research. Other benefits of cloud computing relevant to genomic research include –
- Access to massive storage: Genomic datasets and DNA sequencing information run into several gigabytes for each individual in a species. This enormous data storage requirement balloons the overall costs of genome analytics. However, in line with Kryder’s Law, data storage costs roughly halved every 14 months from 1990 to 2010. Cloud computing has further rendered the storage and efficient processing of voluminous data a cakewalk. Thanks to Cloud’s dynamic scalability, biologists are no longer concerned about potentially running out of disk storage space

Storage cost (Blue) vs. Cost of DNA sequencing (1990-2004: Yellow, 2005-2012: Red):
The introduction of Next Generation Sequencing (NGS) has caused DNA sequencing cost to reduce by half every 5 months. In the future, it will cost less to sequence a DNA than to store it on a hard disk! (Source)
2. Renting reduces budget requirement: In the Cloud model, the Cloud Service Provider (CSP) manages and maintains extensive clusters of computers, storage and high-bandwidth network resources in their data centres. All these utilities, including Graphics Processing Units (GPUs), are offered on a pay-per-use basis at competitive costs. This minimizes the research budget as well as associated maintenance overheads and manpower costs.
3. Seamless scalability: Cloud servers facilitate scalability and reproducibility of genomic analytics across different infrastructure without requiring extensive code refactoring. Furthermore, enormous amounts of data can be stored and processed on the cloud with minimal latency. Researchers can, thus, undertake repeated sequencing when in doubt or subject the same sequence to multiple computational analyses. Healthcare firms can run analytical pipelines across their entire ecosystem instead of relying on deriving actionable insights from piecemeal data.
4. International collaboration: Healthcare/ research firms often cannot find all the genomic information they require from one geographic area. Therefore, these firms collaborate with organizations and researchers across the globe. Cloud-based genome analytics tools support such international collaboration and enable engineers to replicate the research/ re-evaluate the findings.
5. Data security: Protecting genetic and biometric data, including sensitive personal information identifying patients/ volunteers, is critical in genomic/ healthcare research projects. Many nations have brought legislations similar to the Health Insurance Portability and Accountability Act (HIPAA), subjecting information-collecting/ storing organisations to additional compliance regulations. Storing and processing the genomic data on the cloud facilitates data security for information at rest and in transit. Many cloud providers these days are HIPAA-compliant and ensure that the data stored on their servers is never shared with third parties nor accessible to any unauthorized entity. Fascinatingly, this data security comes with cloud computing at no additional cost!

What is the Role of GPU in Genome Analysis?
A single human genome sequence generates approximately 200 GB of raw data. It is projected that over 100 million human genomes will have been sequenced by 2025 as part of various genomic projects. This is equivalent to 20 Exabytes of data. Add another 20 Exabytes for processed data. That’s 40,000,000,000 GB data!
Data compression technologies may somewhat reduce the ginormous storage requirements, but even they are not the complete solution. Furthermore, genome sequencing is meaningless unless followed by thorough analytics and extraction of noteworthy scientific insights. This presupposes the availability of unprecedented computing power at affordable costs. Only Graphics Processing Units (GPUs) fit the bill.
Whereas previously it was uneconomical to envision access to DNA sequencing and genetically customized medication for every individual, unimpeded availability of GPUs and specialized analytics tools via cloud computing servers has upended this.
GPU-accelerated data processing has truly been the harbinger of “genome sequencing revolution”, enabling the crunching of enormous datasets at lightning-fast speeds. Moreover, advancements in GPU technology have also rendered the adoption of cloud computing for genome projects very cost-efficient vis-a-vis in-house infrastructure set-up. The availability of Next-Generation DNA/ RNA Sequencing (NGS) technology and software suites such as Nvidia’s Clara Parabricks has led to genome analytics being performed at incredibly fast speeds.
Vis-a-vis CPU cluster farms delivering equivalent processing power, Cloud GPUs deliver numerous benefits such as –
- Cost efficiency
- Dynamic scalability
- Flexible project pipelining
- Enhanced accuracy
- Integration with various off-the-shelf sequencing suites
Technology Showcase: Nvidia Clara
Nvidia Clara is one of the most popular genome sequencing software suite. Used for numerous common genome analytics tasks, it can analyze 30 human genomes in 25 minutes, instead of 30 hours taken by CPU-based tools! It achieves this through easy integration of GPU- and CPU-driven tasks, utilizing the tremendous computation power offered by GPU’s numerous parallel processing cores to transform the voluminous raw data according to user requirements. Clara users, which include genetic engineers and computational biologists, can thus take advantage of GPU-acceleration across all steps of their analytics workflow.
The Clara Parabricks suite is available as a public container on Nvidia’s NGC portal for use on-prem or on any cloud service. It can be used with Nvidia Ampere series A10, A30, A40, A100 and A6000 GPUs. A 2x GPU set-up running the Clara suite must be supported by a CPU with at least 32 threads and 196 GB RAM. should have at least 100GB CPU RAM and at least 24 CPU threads. Lastly, Clara Parabricks cannot be run in vGPU or MIG environment.
Conclusion
Healthcare firms and gene research institutes are inclining towards GPU computing to enhance their analytics performance. GPU acceleration simultaneously addresses the challenge of high-density data processing while also facilitating scalability and real-time availability. The journey from setbacks to seamless genomic analysis has become possible only because of GPUs and Cloud computing. The day is not far when every human would have access to their genetic data for medical purposes. And what a day it shall be!













